Antimicrobial Resistance Mechanisms
BY KLAUS D. LINSE on JUNE 6, 2017 • ( 0 )
21 Votes
Gram-negative pathogens, such as Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii are now responsible for many infections, including infections occurring in hospitals. Because of their broad spectrum of antibiotic resistance many antibiotics may now longer work well. A few years after the introduction of penicillin, the occurrence of microorganisms able to metabolize this beta-lactam antibiotic was noticed. This phenomenon illustrates that pathogenic microorganisms can develop beta-lactam resistance.
Antimicrobial resistance is now classified as one of the most serious health threats all over the world. Antibiotic resistance can occur in both gram-positive and gram-negative bacteria. However, mechanisms of resistance are more complicated in gram-negative organism. For example, Pseudomonas aeruginosa, Klebsilla pneumonia, and Acinitobacter baumannii tend to be highly resistant to antimicrobial agents. These bacteria can become resistant to almost all antibiotics. Many of these bacteria are resistant to multiple drugs. Therefore they are now known as multi-drug resistant bacteria (MDR GNB).
The use of antimicrobial agents in the food industry is thought to be a major cause for selecting antimicrobial resistance in bacteria. Also, in hospital settings, it is estimated that up to 50% of all antibiotics are needlessly or improperly prescribed. In the US alone, 80% of all antimicrobials are used in the food industry. According to the Center for Disease Control and Prevention (CDC), more than 2 million people are infected with antibiotic-resistant organisms each year. These patients are at increased health risk. Antimicrobial resistance is now classified as one of the most serious health threats worldwide. Furthermore, patients in ICU often become colonized with MDR organisms.
New drugs that circumvent MDR are needed. Recently, a new type of BNA/DNA-peptide conjugate has been developed to treat drug-resistant infections. Lopes et al. in 2015. reported that a BNANC-DNA hybrid co-oligomer conjugated to a cell permeable peptide mimic named CPPBD4 was tested for its ability to reduce the level of resistance in A. baumannii A 155 cells. This compound is now thought to become a useful antimicrobial and chemotherapeutic agent.
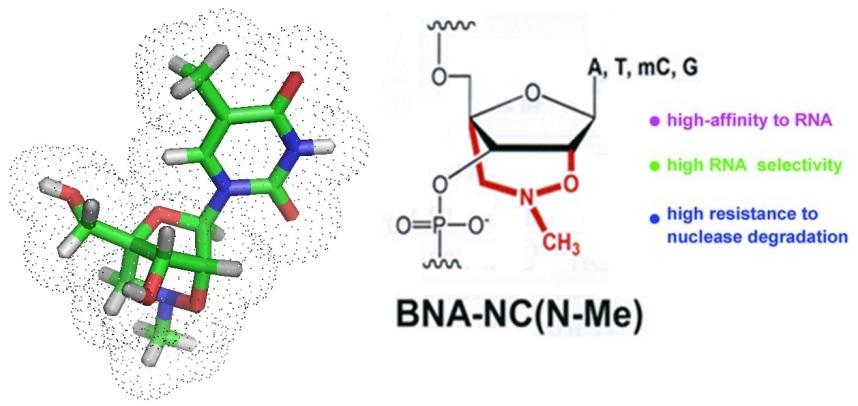
To allow for the detection and identification of bacterial pathogens in blood or tissue new strategies are continuously investigated. Current diagnostic tools are limited in sensitivity, some take a long time to get results, others may not work due to compounds present in the sample analyzed that inhibit PCR, and the occurrence of noncultural microorganisms. Modified nucleic acids such as bridged nucleic acids or BNAs offer themselfs for the design of new, more sensitive diagnositc tools.
Multidrug resistance
Multidrug resistance is defined as resistance by the organism to more than one (1) agent from three (3) or more antimicrobial categories.
Extreme drug resistance
Extreme drug resistance is defined as resistance by the organism to more than one (1) agent in all but two (2) categories.
Pan drug resistance
Pan drug resistance is defined as resistance by the organism to all antibiotics.
Bacterial resistance can occur in a variety of ways as illustrated in Table 1.
Table 1: Mechanisms of Resistance in Gram-negative and Gram-positive Bacteria
| Entero-bacteriaceae | Pseudomonas aeruginosa | Actinobacter | Staphylococcus aureus | |
| AmpC beta-lactamase | √ | √ | √ | |
| ESBL | √ | |||
| Carbapenemases | √ | √ | √ | |
| DNA gyrase / topoisomerase mutations | √ | √ | √ | √ |
| Aminoglycoside-modifying enzymes | √ | √ | √ | |
| Multidrug efflux pumps | √ | √ | ||
| Porin mutations | √ | √ | ||
| Altered penicillin-binding protein | √ | √ | ||
| Penicillinase | √ |
ESBL = extended-spectrum beta-lactamase.
Beta-lactamases
The production of beta-lactamases is a common mechanism of antimicrobial resistance. Beta-lactamases are enzymes that hydrolyze the beta-lactam ring of penicillins, cephalosporins, and carbapenems. Hydrolyzation of the lactam ring renders the drugs inactive. Clinically important beta-lactamases include AmpC, extended-spectrum bata-lactamases (ESBLs) and carbapenemases. ESBLs can hydrolyze penicillins, cephalosporins, and monobactams. Gram-negative pathogens can produce beta-lactamase which is the main factor for acquired beta-lactam resistance.
Four classes of beta-lactamases have been described so far:
A, B, C, and D.
Classes A, C, and D are enzymes with a serine moiety in the active center. These catalyze the hydrolysis of the beta-lactam ring through an acyl-intermediate of serine.
Class B enzymes require a metal cofactor such as zinc in the natural form to function. They are referred to as metallo-beta-lactamases (MBLs).
Treatment of beta-lactamase-mediated resistance
To defeat beta-lactamase-mediated resistance a combination of beta-lactam and a beta-lactamase inhibitor is widely used in the treatment of human infections.
Most of the inhibitors act against class A beta-lactamases but are ineffective against class B, C, and D beta-lactamases.
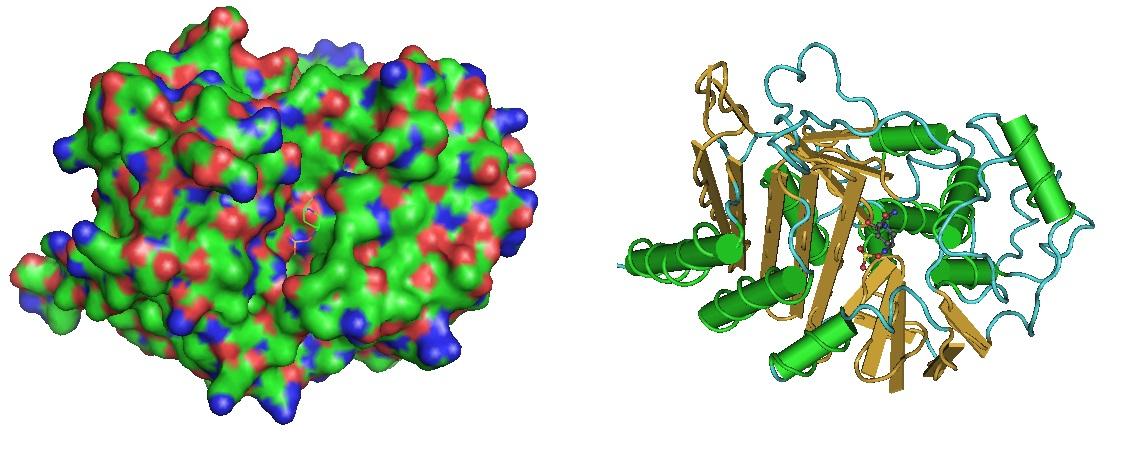 Figure 1: Beta-Lactamase in complex with inhibitor. The structure was reported by Lahiri et al. in 2014. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4187909/, https://www.ncbi.nlm.nih.gov/pubmed/25022578).
Figure 1: Beta-Lactamase in complex with inhibitor. The structure was reported by Lahiri et al. in 2014. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4187909/, https://www.ncbi.nlm.nih.gov/pubmed/25022578).
Mcr-1 gene
The mcr-1 gene confers resistance to polymyxins, including colistin, first described in China, has now been found in bacteria in both humans and meat from animals on five continents, including North America. The mobilized colistin resistance (mcr-1) gene is the reason for plasmid-mediated resistance to colistin. This gene can be transferred horizontally between different bacterial strains. The Mcr gene codes for a phosphatidylethanolamine transferase which transfers a phosphoethanolamine residue to the lipid A present in the cell membrane of gram-negative bacteria. The modified lipid A has a much lower affinity to colistin and other polymyxins. This modification results in reduced activity of the antimicrobial agent.
 Figure 2: Structural models of Phospatidylethanolamine transferase Mcr-1. The mcr-1 gene was identified as a plasmid-mediated resistance mechanism in human and animal Enterobacteriaceae. mcr-1 encodes a membrane-bound enzyme catalysing phosphoethanolamine transfer onto bacterial lipid A. The crystal structure was solved by Hinchliffe et al. in 2017. https://www.ncbi.nlm.nih.gov/pubmed/28059088.
Figure 2: Structural models of Phospatidylethanolamine transferase Mcr-1. The mcr-1 gene was identified as a plasmid-mediated resistance mechanism in human and animal Enterobacteriaceae. mcr-1 encodes a membrane-bound enzyme catalysing phosphoethanolamine transfer onto bacterial lipid A. The crystal structure was solved by Hinchliffe et al. in 2017. https://www.ncbi.nlm.nih.gov/pubmed/28059088.
Major mechanisms of resistance in MDR organisms are illustrated in Table 2. The recommended treatment of MDR organisms begin with broad-spectrum antibiotics for the suspected organism. However the effectiveness of treatment depends on the site of infection, control of the source of infection, and type of resistant organism.
Antimicrobial resistance is a problem requiring a multifaceted approach. The following may be needed to achive success: reduction in total use, improved sanitation and infection control, increased use of rapid diagnostic tests and appropriate vaccines, as well as the development of newer drugs.
Table 2: Recommended Treatment for Multi-Drug Resistant (MDR) Organisms
| Type of Resistance | Common Organism | Recommended Treatment | Comments |
| AmpC beta-lactamse | Enterobacter cloacae; Enterobacteriaceae | Any carbapenem or cefepime | TMP-SMX, quiniolone; tigecycline (Tygacil, Pfizer) also may be effective! |
| ESBL | Klebsiella pneumonia and other Enterobacteriaceae | Any carbapenem | TMP-SMX, quiniolone; tigecycline also may be effective! |
| Carbapenemase | K. pneumonia and other Enterobacteriaceae | Ceftazidime-avibactam (Avycaz, Allergan) ± aztreonam (Azactam, Bristol-Myers Squibb) | Meropenem or tigecycline + polymyxin E (colistin). |
| Alteration of penicillin-binding protein | MRSA | Vancomycin | Daptomycin, telavancin (Vibativ, Theravance), linezolid, TMP-SMX, and ceftaroline (Teflaro, Allergan) are alternatives. |
| Mutation of DNA gyrase and topoisomerase | Enterococcus faecium (VRE) | Linezolid (Zyvox, Pfizer) or daptomycin | Tigecycline may be effective against certain strains of A. baumannii. |
| Decreased permeability plus increased efflux plus de-repressed AmpC | Pseudomonas aeruginosa, Acinetobacter baumanii | Ceftolozane-tazobactum (Zerbxa, Merck) | Tigecycline may be effective against certain strains of A. bumanii. |
| Aminoglycoside-modifying enzymes | P. aeruginosa, A. baumannii | Meropenem (Merrem I.V., AstraZeneca), imipenem-cilastatin (Primaxin I.V., Merck), piperalcillin-tazobactam, or cefepime |
ESBL, extended-spectrum beta-lactamse; MDR, multidrug-resistance; MRSA, methicillin-resistant Staphylococcus aureus; TMP-SMX, trimethoprim-sulfamethoxazole; VRE, vancomycin-resistant enterococci.
Reference
Paul P. Cook; An overview of the current clinical challenges and approaches to patient care. Infectious Disease Special Edition. Spring 2017 59-64.
http://www.nursingworld.org/MainMenuCategories/ANAMarketplace/ANAPeriodicals/OJIN/TableofContents/Vol-16-2011/No2-May-2011/Overview-and-Summary-Patient-Centered-Care.html
http://www.biosyn.com/literaturevault/Inhibition-of-AAC(6%E2%80%B2)-Ib-Mediated-Resistance-to-Amikacin-in-Acinetobacter-baumannii.aspx
https://moh-it.pure.elsevier.com/en/publications/new-beta-lactamases-a-paradigm-for-the-rapid-response-of-bacteria
https://www.ncbi.nlm.nih.gov/pubmed/19747143 Beta-Lactamses
http://jcm.asm.org/content/48/12/4432.full Molecular Probes
—….—
Share this:
-
‹ Teixobactin, a new antibiotic peptide that has the potential to avoid the development of resistance
Categories: Antimicrobial peptide, Bioanalysis, BNAs, Enzymes, Food, Oligonucleotide Synthesis, Peptide Synthesis, peptides, Uncategorized
Tags: Bacteria, Diagnostic Tools, Microorganisms, Multi Drug Resistance, Oligo Synthesis, pathogens, RNA